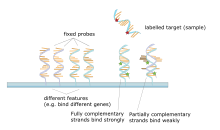Molecular biology
| Part of a series on |
| Biochemistry |
|---|
 |
| Key components |
| History and topics |
| Glossaries |
| Portals: Biology, MCB |
Molecular biology /məˈlɛkjʊlər/ is a branch of biology that concerns the molecular basis of biological activity between biomolecules in the various systems of a cell, including the interactions between DNA, RNA, proteins and their biosynthesis, as well as the regulation of these interactions.[1] Writing in Nature in 1961, William Astbury described molecular biology as:
...not so much a technique as an approach, an approach from the viewpoint of the so-called basic sciences with the leading idea of searching below the large-scale manifestations of classical biology for the corresponding molecular plan. It is concerned particularly with the forms of biological molecules and [...] is predominantly three-dimensional and structural – which does not mean, however, that it is merely a refinement of morphology. It must at the same time inquire into genesis and function.[2]
Contents
Relationship to other biological sciences[edit]
Researchers in molecular biology use specific techniques native to molecular biology but increasingly combine these with techniques and ideas from genetics and biochemistry. There is not a defined line between these disciplines. This is shown in the following schematic that depicts one possible view of the relationships between the fields:[3]
- Biochemistry is the study of the chemical substances and vital processes occurring in live organisms. Biochemists focus heavily on the role, function, and structure of biomolecules. The study of the chemistry behind biological processes and the synthesis of biologically active molecules are examples of biochemistry.[4]
- Genetics is the study of the effect of genetic differences in organisms. This can often be inferred by the absence of a normal component (e.g. one gene). The study of "mutants" – organisms which lack one or more functional components with respect to the so-called "wild type" or normal phenotype. Genetic interactions (epistasis) can often confound simple interpretations of such "knockout" studies.[5]
- Molecular biology is the study of molecular underpinnings of the processes of replication, transcription, translation, and cell function. The central dogma of molecular biology where genetic material is transcribed into RNA and then translated into protein, despite being oversimplified, still provides a good starting point for understanding the field. The picture has been revised in light of emerging novel roles for RNA.[1]
Much of molecular biology is quantitative, and recently much work has been done at its interface with computer science in bioinformatics and computational biology. In the early 2000s, the study of gene structure and function, molecular genetics, has been among the most prominent sub-fields of molecular biology. Increasingly many other areas of biology focus on molecules, either directly studying interactions in their own right such as in cell biology and developmental biology, or indirectly, where molecular techniques are used to infer historical attributes of populations or species, as in fields in evolutionary biology such as population genetics and phylogenetics. There is also a long tradition of studying biomolecules "from the ground up" in biophysics.[citation needed]
Techniques of molecular biology[edit]
Molecular cloning[edit]
One of the most basic techniques of molecular biology to study protein function is molecular cloning. In this technique, DNA coding for a protein of interest is cloned using polymerase chain reaction (PCR), and/or restriction enzymes into a plasmid (expression vector). A vector has 3 distinctive features: an origin of replication, a multiple cloning site (MCS), and a selective marker usually antibiotic resistance. Located upstream of the multiple cloning site are the promoter regions and the transcription start site which regulate the expression of cloned gene. This plasmid can be inserted into either bacterial or animal cells. Introducing DNA into bacterial cells can be done by transformation via uptake of naked DNA, conjugation via cell-cell contact or by transduction via viral vector. Introducing DNA into eukaryotic cells, such as animal cells, by physical or chemical means is called transfection. Several different transfection techniques are available, such as calcium phosphate transfection, electroporation, microinjection and liposome transfection. The plasmid may be integrated into the genome, resulting in a stable transfection, or may remain independent of the genome, called transient transfection.[6][7]
DNA coding for a protein of interest is now inside a cell, and the protein can now be expressed. A variety of systems, such as inducible promoters and specific cell-signaling factors, are available to help express the protein of interest at high levels. Large quantities of a protein can then be extracted from the bacterial or eukaryotic cell. The protein can be tested for enzymatic activity under a variety of situations, the protein may be crystallized so its tertiary structure can be studied, or, in the pharmaceutical industry, the activity of new drugs against the protein can be studied.[citation needed]
Polymerase chain reaction[edit]
Polymerase chain reaction (PCR) is an extremely versatile technique for copying DNA. In brief, PCR allows a specific DNA sequence to be copied or modified in predetermined ways. The reaction is extremely powerful and under perfect conditions could amplify one DNA molecule to become 1.07 billion molecules in less than two hours. The PCR technique can be used to introduce restriction enzyme sites to ends of DNA molecules, or to mutate particular bases of DNA, the latter is a method referred to as site-directed mutagenesis. PCR can also be used to determine whether a particular DNA fragment is found in a cDNA library. PCR has many variations, like reverse transcription PCR (RT-PCR) for amplification of RNA, and, more recently, quantitative PCR which allow for quantitative measurement of DNA or RNA molecules.[8][9]
Gel electrophoresis[edit]
Gel electrophoresis is one of the principal tools of molecular biology. The basic principle is that DNA, RNA, and proteins can all be separated by means of an electric field and size. In agarose gel electrophoresis, DNA and RNA can be separated on the basis of size by running the DNA through an electrically charged agarose gel. Proteins can be separated on the basis of size by using an SDS-PAGE gel, or on the basis of size and their electric charge by using what is known as a 2D gel electrophoresis.[10]
Macromolecule blotting and probing[edit]
The terms northern, western and eastern blotting are derived from what initially was a molecular biology joke that played on the term Southern blotting, after the technique described by Edwin Southern for the hybridisation of blotted DNA. Patricia Thomas, developer of the RNA blot which then became known as the northern blot, actually didn't use the term.[11]
Southern blotting[edit]
Named after its inventor, biologist Edwin Southern, the Southern blot is a method for probing for the presence of a specific DNA sequence within a DNA sample. DNA samples before or after restriction enzyme (restriction endonuclease) digestion are separated by gel electrophoresis and then transferred to a membrane by blotting via capillary action. The membrane is then exposed to a labeled DNA probe that has a complement base sequence to the sequence on the DNA of interest.[12] Southern blotting is less commonly used in laboratory science due to the capacity of other techniques, such as PCR, to detect specific DNA sequences from DNA samples. These blots are still used for some applications, however, such as measuring transgene copy number in transgenic mice or in the engineering of gene knockout embryonic stem cell lines.[citation needed]
Northern blotting[edit]
The northern blot is used to study the expression patterns of a specific type of RNA molecule as relative comparison among a set of different samples of RNA. It is essentially a combination of denaturing RNA gel electrophoresis, and a blot. In this process RNA is separated based on size and is then transferred to a membrane that is then probed with a labeled complement of a sequence of interest. The results may be visualized through a variety of ways depending on the label used; however, most result in the revelation of bands representing the sizes of the RNA detected in sample. The intensity of these bands is related to the amount of the target RNA in the samples analyzed. The procedure is commonly used to study when and how much gene expression is occurring by measuring how much of that RNA is present in different samples. It is one of the most basic tools for determining at what time, and under what conditions, certain genes are expressed in living tissues.[13][14]
Western blotting[edit]
In western blotting, proteins are first separated by size, in a thin gel sandwiched between two glass plates in a technique known as SDS-PAGE. The proteins in the gel are then transferred to a polyvinylidene fluoride (PVDF), nitrocellulose, nylon, or other support membrane. This membrane can then be probed with solutions of antibodies. Antibodies that specifically bind to the protein of interest can then be visualized by a variety of techniques, including colored products, chemiluminescence, or autoradiography. Often, the antibodies are labeled with enzymes. When a chemiluminescent substrate is exposed to the enzyme it allows detection. Using western blotting techniques allows not only detection but also quantitative analysis. Analogous methods to western blotting can be used to directly stain specific proteins in live cells or tissue sections.[15][16]
Eastern blotting[edit]
The eastern blotting technique is used to detect post-translational modification of proteins. Proteins blotted on to the PVDF or nitrocellulose membrane are probed for modifications using specific substrates.[17]
Microarrays[edit]
A DNA microarray is a collection of spots attached to a solid support such as a microscope slide where each spot contains one or more single-stranded DNA oligonucleotide fragments. Arrays make it possible to put down large quantities of very small (100 micrometre diameter) spots on a single slide. Each spot has a DNA fragment molecule that is complementary to a single DNA sequence. A variation of this technique allows the gene expression of an organism at a particular stage in development to be qualified (expression profiling). In this technique the RNA in a tissue is isolated and converted to labeled complementary DNA (cDNA). This cDNA is then hybridized to the fragments on the array and visualization of the hybridization can be done. Since multiple arrays can be made with exactly the same position of fragments they are particularly useful for comparing the gene expression of two different tissues, such as a healthy and cancerous tissue. Also, one can measure what genes are expressed and how that expression changes with time or with other factors. There are many different ways to fabricate microarrays; the most common are silicon chips, microscope slides with spots of ~100 micrometre diameter, custom arrays, and arrays with larger spots on porous membranes (macroarrays). There can be anywhere from 100 spots to more than 10,000 on a given array. Arrays can also be made with molecules other than DNA.[18][19][20][21]
Allele-specific oligonucleotide[edit]
Allele-specific oligonucleotide (ASO) is a technique that allows detection of single base mutations without the need for PCR or gel electrophoresis. Short (20–25 nucleotides in length), labeled probes are exposed to the non-fragmented target DNA, hybridization occurs with high specificity due to the short length of the probes and even a single base change will hinder hybridization. The target DNA is then washed and the labeled probes that didn't hybridize are removed. The target DNA is then analyzed for the presence of the probe via radioactivity or fluorescence. In this experiment, as in most molecular biology techniques, a control must be used to ensure successful experimentation.[22][23]
In molecular biology, procedures and technologies are continually being developed and older technologies abandoned. For example, before the advent of DNA gel electrophoresis (agarose or polyacrylamide), the size of DNA molecules was typically determined by rate sedimentation in sucrose gradients, a slow and labor-intensive technique requiring expensive instrumentation; prior to sucrose gradients, viscometry was used. Aside from their historical interest, it is often worth knowing about older technology, as it is occasionally useful to solve another new problem for which the newer technique is inappropriate.[citation needed]
History[edit]
While molecular biology was established in the 1930s, the term was coined by Warren Weaver in 1938. Weaver was the director of Natural Sciences for the Rockefeller Foundation at the time and believed that biology was about to undergo a period of significant change given recent advances in fields such as X-ray crystallography.[24][25]
Clinical research and medical therapies arising from molecular biology are partly covered under gene therapy. The use of molecular biology or molecular cell biology approaches in medicine is now called molecular medicine. Molecular biology also plays important role in understanding formations, actions, and regulations of various parts of cells which can be used to efficiently target new drugs, diagnosis disease, and understand the physiology of the cell.[26]
See also[edit]
References[edit]
- ^ a b Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Morgan, David; Raff, Martin; Roberts, Keith; Walter, Peter (2014). Molecular Biology of the Cell, Sixth Edition. Garland Science. pp. 1–10. ISBN 978-1-317-56375-4.
- ^ Astbury, W.T. (1961). "Molecular Biology or Ultrastructural Biology?" (PDF). Nature. 190 (4781): 1124. Bibcode:1961Natur.190.1124A. doi:10.1038/1901124a0. PMID 13684868. Retrieved 2008-08-04.
- ^ Lodish, Harvey; Berk, Arnold; Zipursky, S. Lawrence; Matsudaira, Paul; Baltimore, David; Darnell, James (2000). Molecular cell biology (4th ed.). New York: Scientific American Books. ISBN 978-0-7167-3136-8.
- ^ Berg, Jeremy M.; Tymoczko, John L.; Stryer, Lubert; Berg, Jeremy M.; Tymoczko, John L.; Stryer, Lubert (2002). Biochemistry (5th ed.). W H Freeman. ISBN 978-0-7167-3051-4.chapter 1
- ^ Reference, Genetics Home. "Help Me Understand Genetics". Genetics Home Reference. Retrieved 31 December 2016.
- ^ Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter. Isolating, Cloning, and Sequencing DNA. Retrieved 31 December 2016.
- ^ Lessard, Juliane C. (1 January 2013). Molecular cloning. Methods in Enzymology. 529. pp. 85–98. doi:10.1016/B978-0-12-418687-3.00007-0. ISBN 978-0-12-418687-3. ISSN 1557-7988. PMID 24011038. (Subscription required (help)).
- ^ "Polymerase Chain Reaction (PCR)". www.ncbi.nlm.nih.gov. Retrieved 31 December 2016.
- ^ "Polymerase Chain Reaction (PCR) Fact Sheet". National Human Genome Research Institute (NHGRI). Retrieved 31 December 2016.
- ^ Lee, Pei Yun; Costumbrado, John; Hsu, Chih-Yuan; Kim, Yong Hoon (20 April 2012). "Agarose Gel Electrophoresis for the Separation of DNA Fragments". Journal of Visualized Experiments (62). doi:10.3791/3923. ISSN 1940-087X. PMC 4846332. PMID 22546956.
- ^ Thomas, P.S. (1980). "Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose". PNAS. 77 (9): 5201–5205. Bibcode:1980PNAS...77.5201T. doi:10.1073/pnas.77.9.5201. ISSN 1091-6490. PMC 350025. PMID 6159641.
- ^ Brown, T. (1 May 2001). Southern blotting. Current Protocols in Immunology. Chapter 10. pp. Unit 10.6A. doi:10.1002/0471142735.im1006as06. ISBN 978-0-471-14273-7. ISSN 1934-368X. PMID 18432697. (Subscription required (help)).
- ^ Nielsen, Henrik, ed. (2011). RNA methods and protocols. Methods in Molecular Biology. 703. New York: Humana Press. pp. 87–105. doi:10.1007/978-1-59745-248-9_7. ISBN 978-1-59745-248-9. PMID 21125485. (Subscription required (help)).
- ^ He, Shan L. (1 January 2013). Northern blot. Methods in Enzymology. 530. pp. 75–87. doi:10.1016/B978-0-12-420037-1.00003-8. ISBN 978-0-12-420037-1. PMC 4287216. PMID 24034315.
- ^ Mahmood, Tahrin; Yang, Ping-Chang (2016-12-31). "Western Blot: Technique, Theory, and Trouble Shooting". North American Journal of Medical Sciences. 4 (9): 429–434. doi:10.4103/1947-2714.100998. ISSN 2250-1541. PMC 3456489. PMID 23050259.
- ^ Kurien, Biji T.; Scofield, R. Hal (1 April 2006). "Western blotting". Methods. 38 (4): 283–293. doi:10.1016/j.ymeth.2005.11.007. ISSN 1046-2023. PMID 16483794. – via ScienceDirect (Subscription may be required or content may be available in libraries.)
- ^ Thomas, S.; Thirumalapura, N.; Crossley, E. C.; Ismail, N.; Walker, D. H (1 June 2009). "Antigenic protein modifications in Ehrlichia". Parasite Immunology. 31 (6): 296–303. doi:10.1111/j.1365-3024.2009.01099.x. ISSN 1365-3024. PMC 2731653. PMID 19493209.
- ^ "Microarrays". www.ncbi.nlm.nih.gov. Retrieved 31 December 2016.
- ^ Bumgarner, Roger (31 December 2016). Frederick M. Ausubel; et al., eds. "DNA microarrays: Types, Applications and their future". Current protocols in molecular biology. Chapter 22, Unit–22.1. doi:10.1002/0471142727.mb2201s101. ISBN 978-0-471-14272-0. ISSN 1934-3639. PMC 4011503. PMID 23288464.
- ^ Govindarajan, Rajeshwar; Duraiyan, Jeyapradha; Kaliyappan, Karunakaran; Palanisamy, Murugesan (31 December 2016). "Microarray and its applications". Journal of Pharmacy & Bioallied Sciences. 4 (Suppl 2): S310–S312. doi:10.4103/0975-7406.100283. ISSN 0976-4879. PMC 3467903. PMID 23066278.
- ^ Tarca, Adi L.; Romero, Roberto; Draghici, Sorin (31 December 2016). "Analysis of microarray experiments of gene expression profiling". American Journal of Obstetrics and Gynecology. 195 (2): 373–388. doi:10.1016/j.ajog.2006.07.001. ISSN 0002-9378. PMC 2435252. PMID 16890548.
- ^ Cheng, Liang; Zhang, David Y., eds. (2008). Molecular genetic pathology. Totowa, NJ: Humana. p. 96. ISBN 978-1-59745-405-6. Retrieved 31 December 2016.
- ^ Leonard, Debra G.B. (2016). Molecular Pathology in Clinical Practice. Springer. p. 31. ISBN 978-3-319-19674-9. Retrieved 31 December 2016.
- ^ Weaver, Warren (6 November 1970). "Molecular Biology: Origin of the Term". Science. 170 (3958): 581–582. doi:10.1126/science.170.3958.581-a. Retrieved 31 December 2016.
- ^ Bynum, William (1 February 1999). "A History of Molecular Biology". Nature Medicine. 5 (2): 140. doi:10.1038/5498. ISSN 1078-8956. Retrieved 31 December 2016. (Subscription required (help)).
- ^ Bello EA, Schwinn DA. Molecular biology and medicine. A primer for the clinician. Anesthesiology 1996; 85: 1462–1478.
Further reading[edit]
- Cohen, S.N., Chang, A.C.Y., Boyer, H. & Heling, R.B. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. 70, 3240–3244 (1973).
- Rodgers, M. The Pandora's box congress. Rolling Stone 189, 37–77 (1975).
- Keith Roberts, Martin Raff, Bruce Alberts, Peter Walter, Julian Lewis and Alexander Johnson, Molecular Biology of the Cell
- 4th Edition, Routledge, March, 2002, hardcover, 1616 pages, 7.6 pounds, ISBN 0-8153-3218-1
- 3rd Edition, Garland, 1994, ISBN 0-8153-1620-8
- 2nd Edition, Garland, 1989, ISBN 0-8240-3695-6
External links[edit]
 Media related to Molecular biology at Wikimedia Commons
Media related to Molecular biology at Wikimedia Commons- Biochemistry and Molecular Biology at Curlie



.jpg/220px-Two_percent_Agarose_Gel_in_Borate_Buffer_cast_in_a_Gel_Tray_(Front%2C_angled).jpg)