PubMed
 | |
|---|---|
| Contact | |
| Research center | United States National Library of Medicine (NLM) |
| Release date | January 1996 |
| Access | |
| Website | www |
PubMed is a free search engine accessing primarily the MEDLINE database of references and abstracts on life sciences and biomedical topics. The United States National Library of Medicine (NLM) at the National Institutes of Health maintains the database as part of the Entrez system of information retrieval.
From 1971 to 1997, MEDLINE online access to the MEDLARS Online computerized database primarily had been through institutional facilities, such as university libraries. PubMed, first released in January 1996, ushered in the era of private, free, home- and office-based MEDLINE searching.[1] The PubMed system was offered free to the public starting in June 1997.[2]
Contents
- 1 Content
- 2 Characteristics
- 3 Alternative interfaces
- 4 Data mining of PubMed
- 5 See also
- 6 References
- 7 External links
Content[edit]
In addition to MEDLINE, PubMed provides access to:
- older references from the print version of Index Medicus, back to 1951 and earlier
- references to some journals before they were indexed in Index Medicus and MEDLINE, for instance Science, BMJ, and Annals of Surgery
- very recent entries to records for an article before it is indexed with Medical Subject Headings (MeSH) and added to MEDLINE
- a collection of books available full-text and other subsets of NLM records[3]
- PMC citations
- NCBI Bookshelf
Many PubMed records contain links to full text articles, some of which are freely available, often in PubMed Central[4] and local mirrors such as UK PubMed Central.[5]
Information about the journals indexed in MEDLINE, and available through PubMed, is found in the NLM Catalog.[6]
As of 6 December 2018[update], PubMed has more than 29.1 million records going back to 1966, selectively to the year 1865, and very selectively to 1809; about 500,000 new records are added each year. As of the same date[update], 13.1 million of PubMed's records are listed with their abstracts, and 14.2 million articles have links to full-text (of which 3.8 million articles are available, full-text for free for any user).[7] Approximately 12% of the records in PubMed correspond to cancer-related entries, which have grown from 6% in the 1950s to 16% in 2016.[8] Other significant proportion of records correspond to "chemistry" (8.69%), "therapy" (8.39%), and "infection" (5%).[citation needed]
In 2016, NLM changed the indexing system so that publishers will be able to directly correct typos and errors in PubMed indexed articles.[9]
Characteristics[edit]
Standard searches[edit]
Simple searches on PubMed can be carried out by entering key aspects of a subject into PubMed's search window.
PubMed translates this initial search formulation and automatically adds field names, relevant MeSH (Medical Subject Headings) terms, synonyms, Boolean operators, and 'nests' the resulting terms appropriately, enhancing the search formulation significantly, in particular by routinely combining (using the OR operator) textwords and MeSH terms.
The examples given in a PubMed tutorial[10] demonstrate how this automatic process works:
Causes Sleep Walking is translated as ("etiology"[Subheading] OR "etiology"[All Fields] OR "causes"[All Fields] OR "causality"[MeSH Terms] OR "causality"[All Fields]) AND ("somnambulism"[MeSH Terms] OR "somnambulism"[All Fields] OR ("sleep"[All Fields] AND "walking"[All Fields]) OR "sleep walking"[All Fields])
Likewise,
Heart Attack Aspirin Prevention is translated as ("myocardial infarction"[MeSH Terms] OR ("myocardial"[All Fields] AND "infarction"[All Fields]) OR "myocardial infarction"[All Fields] OR ("heart"[All Fields] AND "attack"[All Fields]) OR "heart attack"[All Fields]) AND ("aspirin"[MeSH Terms] OR "aspirin"[All Fields]) AND ("prevention and control"[Subheading] OR ("prevention"[All Fields] AND "control"[All Fields]) OR "prevention and control"[All Fields] OR "prevention"[All Fields])
A new PubMed interface was launched in October 2009 and encouraged the use of such quick, Google-like search formulations; they have also been described as 'telegram' searches.[11] By default the results are sorted by Most Recent, but this can be changed to Best Match, Publication Date, First Author, Last Author, Journal, or Title.[12]
Comprehensive searches[edit]
For optimal searches in PubMed, it is necessary to understand its core component, MEDLINE, and especially of the MeSH (Medical Subject Headings) controlled vocabulary used to index MEDLINE articles. They may also require complex search strategies, use of field names (tags), proper use of limits and other features; reference librarians and search specialists offer search services.[13][14]
Journal article parameters[edit]
When a journal article is indexed, numerous article parameters are extracted and stored as structured information. Such parameters are: Article Type (MeSH terms, e.g., "Clinical Trial"), Secondary identifiers, (MeSH terms), Language, Country of the Journal or publication history (e-publication date, print journal publication date).
Publication Type: Clinical queries/systematic reviews[edit]
Publication type parameter allows searching by the type of publication, including reports of various kinds of clinical research.[15]
Secondary ID[edit]
Since July 2005, the MEDLINE article indexing process extracts identifiers from the article abstract and puts those in a field called Secondary Identifier (SI). The secondary identifier field is to store accession numbers to various databases of molecular sequence data, gene expression or chemical compounds and clinical trial IDs. For clinical trials, PubMed extracts trial IDs for the two largest trial registries: ClinicalTrials.gov (NCT identifier) and the International Standard Randomized Controlled Trial Number Register (IRCTN identifier).[16]
See also[edit]
A reference which is judged particularly relevant can be marked and "related articles" can be identified. If relevant, several studies can be selected and related articles to all of them can be generated (on PubMed or any of the other NCBI Entrez databases) using the 'Find related data' option. The related articles are then listed in order of "relatedness". To create these lists of related articles, PubMed compares words from the title and abstract of each citation, as well as the MeSH headings assigned, using a powerful word-weighted algorithm.[17] The 'related articles' function has been judged to be so precise that the authors of a paper suggested it can be used instead of a full search.[18]
Mapping to MeSH[edit]
PubMed automatically links to MeSH terms and subheadings. Examples would be: "bad breath" links to (and includes in the search) "halitosis", "heart attack" to "myocardial infarction", "breast cancer" to "breast neoplasms". Where appropriate, these MeSH terms are automatically "expanded", that is, include more specific terms. Terms like "nursing" are automatically linked to "Nursing [MeSH]" or "Nursing [Subheading]". This feature is called Auto Term Mapping and is enacted, by default, in free text searching but not exact phrase searching (i.e. enclosing the search query with double quotes).[19] This feature makes PubMed searches more sensitive and avoids false-negative (missed) hits by compensating for the diversity of medical terminology.[19]
My NCBI[edit]
The PubMed optional facility "My NCBI" (with free registration) provides tools for
- saving searches
- filtering search results
- setting up automatic updates sent by e-mail
- saving sets of references retrieved as part of a PubMed search
- configuring display formats or highlighting search terms
and a wide range of other options.[20] The "My NCBI" area can be accessed from any computer with web-access. An earlier version of "My NCBI" was called "PubMed Cubby".[21]
LinkOut[edit]
LinkOut, a NLM facility to link (and make available full-text) local journal holdings.[22] Some 3,200 sites (mainly academic institutions) participate in this NLM facility (as of March 2010[update]), from Aalborg University in Denmark to ZymoGenetics in Seattle.[23] Users at these institutions see their institution's logo within the PubMed search result (if the journal is held at that institution) and can access the full-text.
PubMed Commons[edit]
In 2016, PubMed allows authors of articles to comment on articles indexed by PubMed. This feature was initially tested in a pilot mode (since 2013) and was made permanent in 2016.[24] In February 2018, PubMed Commons was discontinued due to the fact that "usage has remained minimal".[25][26]
PubMed for handhelds/mobiles[edit]
PubMed/MEDLINE can be accessed via handheld devices, using for instance the "PICO" option (for focused clinical questions) created by the NLM.[27] A "PubMed Mobile" option, providing access to a mobile friendly, simplified PubMed version, is also available.[28]
askMEDLINE[edit]
askMEDLINE, a free-text, natural language query tool for MEDLINE/PubMed, developed by the NLM, also suitable for handhelds.[29]
PubMed identifier[edit]
A PMID (PubMed identifier or PubMed unique identifier)[30] is a unique integer value, starting at 1, assigned to each PubMed record. A PMID is not the same as a PMCID which is the identifier for all works published in the free-to-access PubMed Central.[31]
The assignment of a PMID or PMCID to a publication tells the reader nothing about the type or quality of the content. PMIDs are assigned to letters to the editor, editorial opinions, op-ed columns, and any other piece that the editor chooses to include in the journal, as well as peer-reviewed papers. The existence of the identification number is also not proof that the papers have not been retracted for fraud, incompetence, or misconduct. The announcement about any corrections to original papers may be assigned a PMID.
Alternative interfaces[edit]
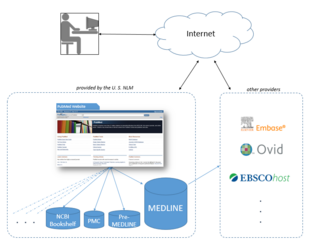
The National Library of Medicine leases the MEDLINE information to a number of private vendors such as Embase, Ovid, Dialog, EBSCO, Knowledge Finder and many other commercial, non-commercial, and academic providers.[32] As of October 2008[update], more than 500 licenses had been issued, more than 200 of them to providers outside the United States. As licenses to use MEDLINE data are available for free, the NLM in effect provides a free testing ground for a wide range[33] of alternative interfaces and 3rd party additions to PubMed, one of a very few large, professionally curated databases which offers this option.
Lu[33] identifies a sample of 28 current and free Web-based PubMed versions, requiring no installation or registration, which are grouped into four categories:
- Ranking search results, for instance: eTBLAST; MedlineRanker;[34] MiSearch;[35]
- Clustering results by topics, authors, journals etc., for instance: Anne O'Tate;[36] ClusterMed;[37]
- Enhancing semantics and visualization, for instance: EBIMed;[38] MedEvi.[39]
- Improved search interface and retrieval experience, for instance, askMEDLINE[40][41] BabelMeSH;[42] and PubCrawler.[43]
As most of these and other alternatives rely essentially on PubMed/MEDLINE data leased under license from the NLM/PubMed, the term "PubMed derivatives" has been suggested.[33] Without the need to store about 90 GB of original PubMed Datasets, anybody can write PubMed applications using the eutils-application program interface as described in "The E-utilities In-Depth: Parameters, Syntax and More", by Eric Sayers, PhD.[44] Various citation format generators, taking PMID numbers as input, are examples of web applications making use of the eutils-application program interface. Sample web pages include Citation Generator - Mick Schroeder, Pubmed Citation Generator - Ultrasound of the Week, PMID2cite, and Cite this for me.
Data mining of PubMed[edit]
Alternative methods to mine the data in PubMed use programming environments such as Matlab, Python or R. In these cases, queries of PubMed are written as lines of code and passed to PubMed and the response is then processed directly in the programming environment. Code can be automated to systematically queries with different keywords such as disease, year, organs, etc. A recent publication (2017) found that the proportion of cancer-related entries in PubMed has risen from 6% in the 1950s to 16% in 2016.[45]
The data accessible by PubMed can be mirrored locally using an unofficial tool such as MEDOC.[46]
See also[edit]
References[edit]
- ^ "PubMed Celebrates its 10th Anniversary". Technical Bulletin. United States National Library of Medicine. 2006-10-05. Retrieved 2011-03-22.
- ^ Lindberg DA (2000). "Internet access to the National Library of Medicine" (PDF). Eff Clin Pract. 3 (5): 256–60. PMID 11185333. Archived from the original (PDF) on 2 November 2013.
- ^ "PubMed: MEDLINE Retrieval on the World Wide Web". Fact Sheet. United States National Library of Medicine. 2002-06-07. Retrieved 2011-03-22.
- ^ Roberts RJ (2001). "PubMed Central: The GenBank of the published literature". Proceedings of the National Academy of Sciences. 98 (2): 381–382. Bibcode:2001PNAS...98..381R. doi:10.1073/pnas.98.2.381. PMC 33354. PMID 11209037.
- ^ McEntyre JR; Ananiadou S; Andrews S; Black WJ; et al. (2010). "UKPMC: A full text article resource for the life sciences". Nucleic Acids Research. 39 (Database issue): D58–D65. doi:10.1093/nar/gkq1063. PMC 3013671. PMID 21062818.
- ^ "NLM Catalogue: Journals referenced in the NCBI Databases". NCBI. 2011.
- ^ (Note: To see the current size of the database simply type "1800:2100[dp]" into the search bar at https://www.ncbi.nlm.nih.gov/pubmed/ and click "search".)
- ^ Reyes-Aldasoro C (2017). "The proportion of cancer-related entries in PubMed has increased considerably; is cancer truly "The Emperor of All Maladies"?". PLOS ONE. 12 (3): e0173671. Bibcode:2017PLoSO..1273671R. doi:10.1371/journal.pone.0173671. PMC 5345838. PMID 28282418.
- ^ "MEDLINE/PubMed Production Improvements Underway".
- ^ "Simple Subject Search with Quiz". NCBI. 2010.
- ^ Clarke J; Wentz R (September 2000). "Pragmatic approach is effective in evidence based health care". BMJ. 321 (7260): 566–567. doi:10.1136/bmj.321.7260.566/a. PMC 1118450. PMID 10968827.
- ^ Fatehi, Farhad; Gray, Leonard C.; Wootton, Richard (January 2014). "How to improve your PubMed/MEDLINE searches: 2. display settings, complex search queries and topic searching". Journal of Telemedicine and Telecare. 20 (1): 44–55. doi:10.1177/1357633X13517067. ISSN 1758-1109. PMID 24352897.
- ^ Jadad AR; McQuay HJ (July 1993). "Searching the literature. Be systematic in your searching". BMJ. 307 (6895): 66. doi:10.1136/bmj.307.6895.66-a. PMC 1678459. PMID 8343701.
- ^ Allison JJ; Kiefe CI; Weissman NW; Carter J; et al. (Spring 1999). "The art and science of searching MEDLINE to answer clinical questions. Finding the right number of articles". Int J Technol Assess Health Care. 15 (2): 281–296. PMID 10507188.
- ^ "Clinical Queries Filter Terms explained". NCBI. 2010.
- ^ Huser V; Cimino JJ (2013). "Evaluating adherence to the International Committee of Medical Journal Editors' policy of mandatory, timely clinical trial registration". J Am Med Inform Assoc. 20 (e1): e169–74. doi:10.1136/amiajnl-2012-001501. PMC 3715364. PMID 23396544.
- ^ "Computation of Related Articles explained". NCBI.
- ^ Chang AA; Heskett KM; Davidson TM (February 2006). "Searching the literature using medical subject headings versus text word with PubMed" (Submitted manuscript). Laryngoscope. 116 (2): 336–340. doi:10.1097/01.mlg.0000195371.72887.a2. PMID 16467730.
- ^ a b Fatehi, Farhad; Gray, Leonard C.; Wootton, Richard (March 2014). "How to improve your PubMed/MEDLINE searches: 3. advanced searching, MeSH and My NCBI". Journal of Telemedicine and Telecare. 20 (2): 102–112. doi:10.1177/1357633X13519036. ISSN 1758-1109. PMID 24614997.
- ^ "My NCBI explained". NCBI. 13 December 2010.
- ^ "PubMed Cubby". Technical Bulletin. United States National Library of Medicine. 2000.
- ^ "LinkOut Overview". NCBI. 2010.
- ^ "LinkOut Participants 2011". NCBI. 2011.
- ^ Team, PubMed Commons (17 December 2015). "Commenting on PubMed: A Successful Pilot".
- ^ "PubMed Commons to be Discontinued". NCBI Insights. 2018-02-01. Retrieved 2018-02-02.
- ^ "PubMed shuts down its comments feature, PubMed Commons". Retraction Watch. 2018-02-02. Retrieved 2018-02-02.
- ^ "PubMed via handhelds (PICO)". Technical Bulletin. United States National Library of Medicine. 2004.
- ^ "PubMed Mobile Beta". Technical Bulletin. United States National Library of Medicine. 2011.
- ^ "askMedline". NCBI. 2005.
- ^ "Search Field Descriptions and Tags". National Center for Biotechnology Information. Retrieved 15 July 2013.
- ^ Keener, Molly (19 May 2008). "PMID vs. PMCID: What's the difference?" (PDF). University of Chicago. Retrieved 19 January 2014.
- ^ "Leasing journal citations from PubMed/Medline". NLM. 2011.
- ^ a b c Lu Z (2011). "PubMed and beyond: A survey of web tools for searching biomedical literature". Database. 2011: baq036. doi:10.1093/database/baq036. PMC 3025693. PMID 21245076.
- ^ Fontaine JF; Barbosa-Silva A; Schaefer M; Huska MR; et al. (2009). "MedlineRanker: Flexible ranking of biomedical literature". Nucleic Acids Research. 37 (Web Server issue): W141–W146. doi:10.1093/nar/gkp353. PMC 2703945. PMID 19429696.
- ^ States DJ; Ade AS; Wright ZC; Bookvich AV; et al. (2009). "MiSearch adaptive pubMed search tool". Bioinformatics. 25 (7): 974–976. doi:10.1093/bioinformatics/btn033. PMC 2660869. PMID 18326507.
- ^ Smalheiser NR; Zhou W; Torvik VI (2008). "Anne O'Tate: A tool to support user-driven summarization, drill-down and browsing of PubMed search results". Journal of Biomedical Discovery and Collaboration. 3: 2. doi:10.1186/1747-5333-3-2. PMC 2276193. PMID 18279519.
- ^ "ClusterMed". Vivisimo Clustering Engine. 2011.
- ^ Rebholz-Schuhmann D; Kirsch H; Arregui M; Gaudan S; et al. (2007). "EBIMed--text crunching to gather facts for proteins from Medline". Bioinformatics. 23 (2): e237–e244. doi:10.1093/bioinformatics/btl302. PMID 17237098.
- ^ Kim JJ; Pezik P; Rebholz-Schuhmann D (2008). "MedEvi: Retrieving textual evidence of relations between biomedical concepts from Medline". Bioinformatics. 24 (11): 1410–1412. doi:10.1093/bioinformatics/btn117. PMC 2387223. PMID 18400773.
- ^ Fontelo P; Liu F; Ackerman M; Schardt CM; et al. (2006). "AskMEDLINE: A report on a year-long experience". AMIA Annual Symposium Proceedings. 2006: 923. PMC 1839379. PMID 17238542.
- ^ Fontelo P; Liu F; Ackerman M (2005). "MeSH Speller + askMEDLINE: Auto-completes MeSH terms then searches MEDLINE/PubMed via free-text, natural language queries". AMIA Annual Symposium Proceedings. 2005: 957. PMC 1513542. PMID 16779244.
- ^ Fontelo P; Liu F; Leon S; Anne A; et al. (2007). "PICO Linguist and BabelMeSH: Development and partial evaluation of evidence-based multilanguage search tools for MEDLINE/PubMed". Studies in Health Technology and Informatics. 129 (Pt 1): 817–821. PMID 17911830.
- ^ Hokamp K; Wolfe KH (2004). "PubCrawler: Keeping up comfortably with PubMed and GenBank". Nucleic Acids Research. 32 (Web Server issue): W16–W19. doi:10.1093/nar/gkh453. PMC 441591. PMID 15215341.
- ^ Eric Sayers, PhD. "The E-utilities In-Depth: Parameters, Syntax and More". NCBI.
- ^ Reyes-Aldasoro C (2017). "The proportion of cancer-related entries in PubMed has increased considerably; is cancer truly "The Emperor of All Maladies"?". PLOS ONE. 12 (3): e0173671. Bibcode:2017PLoSO..1273671R. doi:10.1371/journal.pone.0173671. PMC 5345838. PMID 28282418.
- ^ "MEDOC (MEdline DOwnloading Contrivance)". 2017.
External links[edit]
Wikidata has the property:
|