Longest common substring problem
In computer science, the longest common substring problem is to find the longest string (or strings) that is a substring (or are substrings) of two or more strings.
Contents
Example[edit]
The longest common substring of the strings "ABABC", "BABCA" and "ABCBA" is string "ABC" of length 3. Other common substrings are "A", "AB", "B", "BA", "BC" and "C".
ABABC
|||
BABCA
|||
ABCBA
Problem definition[edit]
Given two strings, of length and of length , find the longest strings which are substrings of both and .
A generalization is the k-common substring problem. Given the set of strings , where and . Find for each , the longest strings which occur as substrings of at least strings.
Algorithms[edit]
One can find the lengths and starting positions of the longest common substrings of and in time with the help of a generalized suffix tree. Finding them by dynamic programming costs . The solutions to the generalized problem take space and ·...· time with dynamic programming and take time with generalized suffix tree.
Suffix tree[edit]
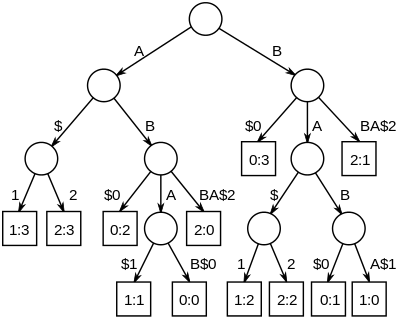
The longest common substrings of a set of strings can be found by building a generalized suffix tree for the strings, and then finding the deepest internal nodes which have leaf nodes from all the strings in the subtree below it. The figure on the right is the suffix tree for the strings "ABAB", "BABA" and "ABBA", padded with unique string terminators, to become "ABAB$0", "BABA$1" and "ABBA$2". The nodes representing "A", "B", "AB" and "BA" all have descendant leaves from all of the strings, numbered 0, 1 and 2.
Building the suffix tree takes time (if the size of the alphabet is constant). If the tree is traversed from the bottom up with a bit vector telling which strings are seen below each node, the k-common substring problem can be solved in time. If the suffix tree is prepared for constant time lowest common ancestor retrieval, it can be solved in time.[1]
Dynamic programming[edit]
First find the longest common suffix for all pairs of prefixes of the strings. The longest common suffix is
For the example strings "ABAB" and "BABA":
| A | B | A | B | ||
|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | |
| B | 0 | 0 | 1 | 0 | 1 |
| A | 0 | 1 | 0 | 2 | 0 |
| B | 0 | 0 | 2 | 0 | 3 |
| A | 0 | 1 | 0 | 3 | 0 |
The maximal of these longest common suffixes of possible prefixes must be the longest common substrings of S and T. These are shown on diagonals, in red, in the table. For this example, the longest common substrings are "BAB" and "ABA".
This can be extended to more than two strings by adding more dimensions to the table.
Pseudocode[edit]
The following pseudocode finds the set of longest common substrings between two strings with dynamic programming:
function LCSubstr(S[1..r], T[1..n])
L := array(1..r, 1..n)
z := 0
ret := {}
for i := 1..r
for j := 1..n
if S[i] == T[j]
if i == 1 or j == 1
L[i,j] := 1
else
L[i,j] := L[i-1,j-1] + 1
if L[i,j] > z
z := L[i,j]
ret := {S[i-z+1..i]}
else
if L[i,j] == z
ret := ret ∪ {S[i-z+1..i]}
else
L[i,j] := 0
return ret
This algorithm runs in time. The variable z is used to hold the length of the longest common substring found so far. The set ret is used to hold the set of strings which are of length z. The set ret can be saved efficiently by just storing the index i, which is the last character of the longest common substring (of size z) instead of S[i-z+1..i]. Thus all the longest common substrings would be, for each i in ret, S[(ret[i]-z)..(ret[i])].
The following tricks can be used to reduce the memory usage of an implementation:
- Keep only the last and current row of the DP table to save memory ( instead of )
- Store only non-zero values in the rows. This can be done using hash tables instead of arrays. This is useful for large alphabets.
See also[edit]
- Longest palindromic substring
- n-gram, all the possible substrings of length n that are contained in a string
References[edit]
- ^ Gusfield, Dan (1999) [1997]. Algorithms on Strings, Trees and Sequences: Computer Science and Computational Biology. USA: Cambridge University Press. ISBN 0-521-58519-8.
External links[edit]
| The Wikibook Algorithm implementation has a page on the topic of: Longest common substring |
- Dictionary of Algorithms and Data Structures: longest common substring
- Perl/XS implementation of the dynamic programming algorithm
- Perl/XS implementation of the suffix tree algorithm
- Dynamic programming implementations in various languages on wikibooks
- working AS3 implementation of the dynamic programming algorithm
- Suffix Tree based C implementation of Longest common substring for two strings

















![{\mathit {LCSuff}}(S_{{1..p}},T_{{1..q}})={\begin{cases}{\mathit {LCSuff}}(S_{{1..p-1}},T_{{1..q-1}})+1&{\mathrm {if}}\;S[p]=T[q]\\0&{\mathrm {otherwise}}.\end{cases}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/83ccdb67e41ba0b5043a8eb2a67ca0d7a6908ad2)


