Self-organizing map
This article may require cleanup to meet Wikipedia's quality standards. (June 2011) (Learn how and when to remove this template message) |
| Machine learning and data mining |
|---|
 |
|
Machine-learning venues |
A self-organizing map (SOM) or self-organizing feature map (SOFM) is a type of artificial neural network (ANN) that is trained using unsupervised learning to produce a low-dimensional (typically two-dimensional), discretized representation of the input space of the training samples, called a map, and is therefore a method to do dimensionality reduction. Self-organizing maps differ from other artificial neural networks as they apply competitive learning as opposed to error-correction learning (such as backpropagation with gradient descent), and in the sense that they use a neighborhood function to preserve the topological properties of the input space.

This makes SOMs useful for visualization by creating low-dimensional views of high-dimensional data, akin to multidimensional scaling. The artificial neural network introduced by the Finnish professor Teuvo Kohonen in the 1980s is sometimes called a Kohonen map or network.[1][2] The Kohonen net is a computationally convenient abstraction building on biological models of neural systems from the 1970s[3] and morphogenesis models dating back to Alan Turing in the 1950s.[4]
While it is typical to consider this type of network structure as related to feedforward networks where the nodes are visualized as being attached, this type of architecture is fundamentally different in arrangement and motivation.
Useful extensions include using toroidal grids where opposite edges are connected and using large numbers of nodes.
It has been shown that while self-organizing maps with a small number of nodes behave in a way that is similar to K-means, larger self-organizing maps rearrange data in a way that is fundamentally topological in character.
It is also common to use the U-Matrix.[5] The U-Matrix value of a particular node is the average distance between the node's weight vector and that of its closest neighbors.[6] In a square grid, for instance, we might consider the closest 4 or 8 nodes (the Von Neumann and Moore neighborhoods, respectively), or six nodes in a hexagonal grid.
Large SOMs display emergent properties. In maps consisting of thousands of nodes, it is possible to perform cluster operations on the map itself.[7]
Contents
Structure and operations[edit]
Like most artificial neural networks, SOMs operate in two modes: training and mapping. "Training" builds the map using input examples (a competitive process, also called vector quantization), while "mapping" automatically classifies a new input vector.
The visible part of a self-organizing map is the map space, which consists of components called nodes or neurons. The map space is defined beforehand, usually as a finite two-dimensional region where nodes are arranged in a regular hexagonal or rectangular grid.[8] Each node is associated with a "weight" vector, which is a position in the input space; that is, it has the same dimension as each input vector. While nodes in the map space stay fixed, training consists in moving weight vectors toward the input data (reducing a distance metric) without spoiling the topology induced from the map space. Thus, the self-organizing map describes a mapping from a higher-dimensional input space to a lower-dimensional map space. Once trained, the map can classify a vector from the input space by finding the node with the closest (smallest distance metric) weight vector to the input space vector.
Learning algorithm[edit]
The goal of learning in the self-organizing map is to cause different parts of the network to respond similarly to certain input patterns. This is partly motivated by how visual, auditory or other sensory information is handled in separate parts of the cerebral cortex in the human brain.[9]
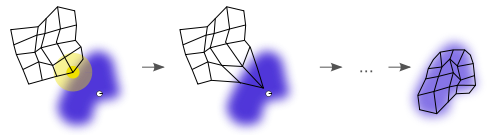
The weights of the neurons are initialized either to small random values or sampled evenly from the subspace spanned by the two largest principal component eigenvectors. With the latter alternative, learning is much faster because the initial weights already give a good approximation of SOM weights.[10]
The network must be fed a large number of example vectors that represent, as close as possible, the kinds of vectors expected during mapping. The examples are usually administered several times as iterations.
The training utilizes competitive learning. When a training example is fed to the network, its Euclidean distance to all weight vectors is computed. The neuron whose weight vector is most similar to the input is called the best matching unit (BMU). The weights of the BMU and neurons close to it in the SOM grid are adjusted towards the input vector. The magnitude of the change decreases with time and with the grid-distance from the BMU. The update formula for a neuron v with weight vector Wv(s) is
- ,
where s is the step index, t an index into the training sample, u is the index of the BMU for D(t), α(s) is a monotonically decreasing learning coefficient and D(t) is the input vector; Θ(u, v, s) is the neighborhood function which gives the distance between the neuron u and the neuron v in step s.[11] Depending on the implementations, t can scan the training data set systematically (t is 0, 1, 2...T-1, then repeat, T being the training sample's size), be randomly drawn from the data set (bootstrap sampling), or implement some other sampling method (such as jackknifing).
The neighborhood function Θ(u, v, s) depends on the grid-distance between the BMU (neuron u) and neuron v. In the simplest form it is 1 for all neurons close enough to BMU and 0 for others, but a Gaussian function is a common choice, too. Regardless of the functional form, the neighborhood function shrinks with time.[9] At the beginning when the neighborhood is broad, the self-organizing takes place on the global scale. When the neighborhood has shrunk to just a couple of neurons, the weights are converging to local estimates. In some implementations the learning coefficient α and the neighborhood function Θ decrease steadily with increasing s, in others (in particular those where t scans the training data set) they decrease in step-wise fashion, once every T steps.
This process is repeated for each input vector for a (usually large) number of cycles λ. The network winds up associating output nodes with groups or patterns in the input data set. If these patterns can be named, the names can be attached to the associated nodes in the trained net.
During mapping, there will be one single winning neuron: the neuron whose weight vector lies closest to the input vector. This can be simply determined by calculating the Euclidean distance between input vector and weight vector.
While representing input data as vectors has been emphasized in this article, it should be noted that any kind of object which can be represented digitally, which has an appropriate distance measure associated with it, and in which the necessary operations for training are possible can be used to construct a self-organizing map. This includes matrices, continuous functions or even other self-organizing maps.
Variables[edit]
These are the variables needed, with vectors in bold,
- is the current iteration
- is the iteration limit
- is the index of the target input data vector in the input data set
- is a target input data vector
- is the index of the node in the map
- is the current weight vector of node
- is the index of the best matching unit (BMU) in the map
- is a restraint due to distance from BMU, usually called the neighborhood function, and
- is a learning restraint due to iteration progress.
Algorithm[edit]
- Randomize the node weight vectors in a map
- Randomly pick an input vector
- Traverse each node in the map
- Use the Euclidean distance formula to find the similarity between the input vector and the map's node's weight vector
- Track the node that produces the smallest distance (this node is the best matching unit, BMU)
- Update the weight vectors of the nodes in the neighborhood of the BMU (including the BMU itself) by pulling them closer to the input vector
- Increase and repeat from step 2 while
A variant algorithm:
- Randomize the map's nodes' weight vectors
- Traverse each input vector in the input data set
- Traverse each node in the map
- Use the Euclidean distance formula to find the similarity between the input vector and the map's node's weight vector
- Track the node that produces the smallest distance (this node is the best matching unit, BMU)
- Update the nodes in the neighborhood of the BMU (including the BMU itself) by pulling them closer to the input vector
- Traverse each node in the map
- Increase and repeat from step 2 while
SOM Initialization[edit]
Selection of a good initial approximation is a well-known problem for all iterative methods of learning neural networks. Kohonen[12] used random initiation of SOM weights. Recently, principal component initialization, in which initial map weights are chosen from the space of the first principal components, has become popular due to the exact reproducibility of the results.[13]
Careful comparison of the random initiation approach to principal component initialization for one-dimensional SOM (models of principal curves) demonstrated that the advantages of principal component SOM initialization are not universal. The best initialization method depends on the geometry of the specific dataset. Principal component initialization is preferable (in dimension one) if the principal curve approximating the dataset can be univalently and linearly projected on the first principal component (quasilinear sets). For nonlinear datasets, however, random initiation performs better.[14]
Examples[edit]
Fisher's Iris Flower Data[edit]
This section does not cite any sources. (February 2010) (Learn how and when to remove this template message) |
This section possibly contains original research. (June 2017) (Learn how and when to remove this template message) |
Consider an n×m array of nodes, each of which contains a weight vector and is aware of its location in the array. Each weight vector is of the same dimension as the node's input vector. The weights may initially be set to random values.
Now we need input to feed the map. Colors can be represented by their red, green, and blue components. Consequently, we will represent colors as vectors in the unit cube of the free vector space over ℝ generated by the basis:
- R = <255, 0, 0>
- G = <0, 255, 0>
- B = <0, 0, 255>
The diagram shown
compares the results of training on the data sets[Note 1]
- threeColors = [255, 0, 0], [0, 255, 0], [0, 0, 255]
- eightColors = [0, 0, 0], [255, 0, 0], [0, 255, 0], [0, 0, 255], [255, 255, 0], [0, 255, 255], [255, 0, 255], [255, 255, 255]
and the original images. Note the striking resemblance between the two.
Similarly, after training a 40×40 grid of neurons for 250 iterations with a learning rate of 0.1 on Fisher's Iris, the map can already detect the main differences between species.

Interpretation[edit]


There are two ways to interpret a SOM. Because in the training phase weights of the whole neighborhood are moved in the same direction, similar items tend to excite adjacent neurons. Therefore, SOM forms a semantic map where similar samples are mapped close together and dissimilar ones apart. This may be visualized by a U-Matrix (Euclidean distance between weight vectors of neighboring cells) of the SOM.[5][6][16]
The other way is to think of neuronal weights as pointers to the input space. They form a discrete approximation of the distribution of training samples. More neurons point to regions with high training sample concentration and fewer where the samples are scarce.
SOM may be considered a nonlinear generalization of Principal components analysis (PCA).[17] It has been shown, using both artificial and real geophysical data, that SOM has many advantages[18][19] over the conventional feature extraction methods such as Empirical Orthogonal Functions (EOF) or PCA.
Originally, SOM was not formulated as a solution to an optimisation problem. Nevertheless, there have been several attempts to modify the definition of SOM and to formulate an optimisation problem which gives similar results.[20] For example, Elastic maps use the mechanical metaphor of elasticity to approximate principal manifolds:[21] the analogy is an elastic membrane and plate.
Alternatives[edit]
- The generative topographic map (GTM) is a potential alternative to SOMs. In the sense that a GTM explicitly requires a smooth and continuous mapping from the input space to the map space, it is topology preserving. However, in a practical sense, this measure of topological preservation is lacking.[22]
- The time adaptive self-organizing map (TASOM) network is an extension of the basic SOM. The TASOM employs adaptive learning rates and neighborhood functions. It also includes a scaling parameter to make the network invariant to scaling, translation and rotation of the input space. The TASOM and its variants have been used in several applications including adaptive clustering, multilevel thresholding, input space approximation, and active contour modeling.[23] Moreover, a Binary Tree TASOM or BTASOM, resembling a binary natural tree having nodes composed of TASOM networks has been proposed where the number of its levels and the number of its nodes are adaptive with its environment.[24]
- The growing self-organizing map (GSOM) is a growing variant of the self-organizing map. The GSOM was developed to address the issue of identifying a suitable map size in the SOM. It starts with a minimal number of nodes (usually four) and grows new nodes on the boundary based on a heuristic. By using a value called the spread factor, the data analyst has the ability to control the growth of the GSOM.
- The elastic maps approach[25] borrows from the spline interpolation the idea of minimization of the elastic energy. In learning, it minimizes the sum of quadratic bending and stretching energy with the least squares approximation error.
- The conformal approach [26][27] that uses conformal mapping to interpolate each training sample between grid nodes in a continuous surface. A one-to-one smooth mapping is possible in this approach.
- The oriented and scalable map (OS-Map) generalises the neighborhood function and the winner selection[28]. The homogeneous Gaussian neighborhood function is replaced with the matrix exponential. Thus one can specify the orientation either in the map space or in the data space. SOM has a fixed scale (=1), so that the maps "optimally describe the domain of observation". But what about a map covering the domain twice or in n-folds? This entails the conception of scaling. The OS-Map regards the scale as a statistical description of how many best-matching nodes an input has in the map.
Applications[edit]
- Meteorology and oceanography[29]
- Project prioritization and selection [30]
- Seismic facies analysis for oil and gas exploration [31]
- Failure mode and effects analysis [32]
- Creation of artwork [33]
See also[edit]
- Neural gas
- Learning Vector Quantization
- Liquid state machine
- Hybrid Kohonen SOM
- Sparse coding
- Sparse distributed memory
- Deep learning
- Neocognitron
- Topological data analysis
Notes[edit]
- ^ These data sets are not normalized. Normalization would be necessary to train the SOM.
References[edit]
- ^ Kohonen, Teuvo; Honkela, Timo (2007). "Kohonen Network". Scholarpedia.
- ^ Kohonen, Teuvo (1982). "Self-Organized Formation of Topologically Correct Feature Maps". Biological Cybernetics. 43 (1): 59–69. doi:10.1007/bf00337288.
- ^ Von der Malsburg, C (1973). "Self-organization of orientation sensitive cells in the striate cortex". Kybernetik. 14 (2): 85–100. doi:10.1007/bf00288907. PMID 4786750.
- ^ Turing, Alan (1952). "The chemical basis of morphogenesis". Phil. Trans. R. Soc. 237: 5–72.
- ^ a b Ultsch, Alfred; Siemon, H. Peter (1990). "Kohonen's Self Organizing Feature Maps for Exploratory Data Analysis". In Widrow, Bernard; Angeniol, Bernard. Proceedings of the International Neural Network Conference (INNC-90), Paris, France, July 9–13, 1990. 1. Dordrecht, Netherlands: Kluwer. pp. 305–308. ISBN 978-0-7923-0831-7. Archived from the original on 2013-06-13.
- ^ a b Ultsch, Alfred (2003); U*-Matrix: A tool to visualize clusters in high dimensional data, Department of Computer Science, University of Marburg, Technical Report Nr. 36:1-12
- ^ Ultsch, Alfred (2007). "Emergence in Self-Organizing Feature Maps". In Ritter, H.; Haschke, R. Proceedings of the 6th International Workshop on Self-Organizing Maps (WSOM '07). Bielefeld, Germany: Neuroinformatics Group. ISBN 978-3-00-022473-7.
- ^ Jaakko Hollmen (9 March 1996). "Self-Organizing Map (SOM)". Aalto University.
- ^ a b Haykin, Simon (1999). "9. Self-organizing maps". Neural networks - A comprehensive foundation (2nd ed.). Prentice-Hall. ISBN 978-0-13-908385-3.
- ^ Kohonen, Teuvo (2005). "Intro to SOM". SOM Toolbox. Retrieved 2006-06-18.
- ^ Kohonen, Teuvo; Honkela, Timo (2011). "Kohonen network". Scholarpedia. Retrieved 2012-09-24.
- ^ T. Kohonen, Self-Organization and Associative Memory. Springer, Berlin, 1984.
- ^ A. Ciampi, Y. Lechevallier, Clustering large, multi-level data sets: An approach based on Kohonen self organizing maps, in D.A. Zighed, J. Komorowski, J. Zytkow (Eds.), PKDD 2000, Springer LNCS (LNAI), vol. 1910, pp. 353-358, 2000.
- ^ Akinduko, A.A.; Mirkes, E.M.; Gorban, A.N. (2016). "SOM: Stochastic initialization versus principal components". Information Sciences. 364-365: 213–221. doi:10.1016/j.ins.2015.10.013.
- ^ Illustration is prepared using free software: Mirkes, Evgeny M.; Principal Component Analysis and Self-Organizing Maps: applet, University of Leicester, 2011
- ^ Saadatdoost, Robab, Alex Tze Hiang Sim, and Jafarkarimi, Hosein. "Application of self organizing map for knowledge discovery based in higher education data." Research and Innovation in Information Systems (ICRIIS), 2011 International Conference on. IEEE, 2011.
- ^ Yin, Hujun; Learning Nonlinear Principal Manifolds by Self-Organising Maps, in Gorban, Alexander N.; Kégl, Balázs; Wunsch, Donald C.; and Zinovyev, Andrei (Eds.); Principal Manifolds for Data Visualization and Dimension Reduction, Lecture Notes in Computer Science and Engineering (LNCSE), vol. 58, Berlin, Germany: Springer, 2008, ISBN 978-3-540-73749-0
- ^ Liu, Yonggang; Weisberg, Robert H (2005). "Patterns of Ocean Current Variability on the West Florida Shelf Using the Self-Organizing Map". Journal of Geophysical Research. 110 (C6): C06003. Bibcode:2005JGRC..110.6003L. doi:10.1029/2004JC002786.
- ^ Liu, Yonggang; Weisberg, Robert H.; Mooers, Christopher N. K. (2006). "Performance Evaluation of the Self-Organizing Map for Feature Extraction". Journal of Geophysical Research. 111 (C5): C05018. Bibcode:2006JGRC..111.5018L. doi:10.1029/2005jc003117.
- ^ Heskes, Tom; Energy Functions for Self-Organizing Maps, in Oja, Erkki; and Kaski, Samuel (Eds.), Kohonen Maps, Elsevier, 1999
- ^ Gorban, Alexander N.; Kégl, Balázs; Wunsch, Donald C.; and Zinovyev, Andrei (Eds.); Principal Manifolds for Data Visualization and Dimension Reduction, Lecture Notes in Computer Science and Engineering (LNCSE), vol. 58, Berlin, Germany: Springer, 2008, ISBN 978-3-540-73749-0
- ^ Kaski, Samuel (1997). Data Exploration Using Self-Organizing Maps. Acta Polytechnica Scandinavica. Mathematics, Computing and Management in Engineering Series No. 82. Espoo, Finland: Finnish Academy of Technology. ISBN 978-952-5148-13-8.
- ^ Shah-Hosseini, Hamed; Safabakhsh, Reza (April 2003). "TASOM: A New Time Adaptive Self-Organizing Map". IEEE Transactions on Systems, Man, and Cybernetics—Part B: Cybernetics. 33 (2): 271–282. doi:10.1109/tsmcb.2003.810442. PMID 18238177.
- ^ Shah-Hosseini, Hamed (May 2011). "Binary Tree Time Adaptive Self-Organizing Map". Neurocomputing. 74 (11): 1823–1839. doi:10.1016/j.neucom.2010.07.037.
- ^ A. N. Gorban, A. Zinovyev, Principal manifolds and graphs in practice: from molecular biology to dynamical systems, International Journal of Neural Systems, Vol. 20, No. 3 (2010) 219–232.
- ^ Liou, C.-Y.; Kuo, Y.-T. (2005). "Conformal Self-organizing Map for a Genus Zero Manifold". The Visual Computer. 21 (5): 340–353. doi:10.1007/s00371-005-0290-6.
- ^ Liou, C.-Y.; Tai, W.-P. (2000). "Conformality in the self-organization network". Artificial Intelligence. 116 (1–2): 265–286. doi:10.1016/S0004-3702(99)00093-4.
- ^ Hua, H., 2016. Image and geometry processing with Oriented and Scalable Map. Neural Networks, 77, pp.1-6.
- ^ Liu, Y., and R.H. Weisberg (2011) A review of self-organizing map applications in meteorology and oceanography. In: Self-Organizing Maps-Applications and Novel Algorithm Design, 253-272.
- ^ Zheng, G. and Vaishnavi, V. (2011) "A Multidimensional Perceptual Map Approach to Project Prioritization and Selection," AIS Transactions on Human-Computer Interaction (3) 2, pp. 82-103
- ^ Taner, M. T.; Walls, J. D.; Smith, M.; Taylor, G.; Carr, M. B.; Dumas, D. (2001). "Reservoir characterization by calibration of self-organized map clusters". SEG Technical Program Expanded Abstracts. 2001: 1552–1555. doi:10.1190/1.1816406.
- ^ Chang, Wui Lee; Pang, Lie Meng; Tay, Kai Meng (March 2017). "Application of Self-Organizing Map to Failure Modes and Effects Analysis Methodology". Neurocomputing. PP: 314–320. doi:10.1016/j.neucom.2016.04.073.
- ^ ANNetGPGPU CUDA Library with examples [1] GPU accelerated image creation
| Wikimedia Commons has media related to Self-organizing map. |












